International Journal of Biology and Nanobiomaterials 1 (2021) 47-66
Original Article
ESBL genes detected in Pseudomonas aeruginosa isolated from dental sources
Vignesh Pillaia, Aneesh Nairb, Soundharya Rb, Florida Tiltonb, Priya M*c
aDepartment of Microbiology, Nehru Arts and Science College, Coimbatore
bBiozone Research Technologies Pvt. Ltd, Chennai
cPSG College of Arts & Science, Coimbatore
ARTICLE INFO
Article history:
Received 20 Jan 2021
Revised 23 Feb 2021
Accepted 4 Mar 2021
Available online 7 Mar 2021
Keywords:
Pseudomonas aeruginosa
Wrightiatinctoria
Quercetin 3-sophorosidePhytocompounds,
Antibiotic
ABSTRACT
Antibiotic resistance is a major crisis during the treatment of nosocomial infections especially in the 20th century. Among some common microbesPseudomonas aeruginosa is seen to spontaneously develop into multidrug resistant strain (MDR). Pseudomonas aeruginosa has developed to be a multidrug resistant strain to show resistance towards many common antibiotics. Both antibiotics and natural sources individually have not been able to inhibit the virulent growth of the microbe because of which the infection persists. An in-silico approach to find out which phytochemical compounds have could inhibit the MexAB efflux pump, has been studied. As, this system that has been a frequently reported reason for antibiotic resistance in Pseudomonas aeruginosa. Ligands present in the plant Wrightiatinctoria, have been reported previously for inhibitory activity against Pseudomonas aeruginosa. These were used to dock with the efflux pump in study. Quercetin 3-sophoroside and quercetin glycoside has shown best pose with this particular receptor. The binding energy was recorded to be -141.93J. In this report, a resistant strain of P. aeruginosaisolated from dental wounds is targeted to find out the possible mechanism of resistance present in it. The research could target two such mechanisms- efflux pump activity and resistive genes.
- Introduction
For the past 20 years, continuous changes in infection control practises, health care and antimicrobial use during resistance is said to have influenced the frequency of Gram-negative organisms to be associated with nosocomial infections. The most frequently reported types include- Surgical site infection (SSI), Urinary Tract Infections (UTI) and Bloodstream Infections (BSI). Research in 2003 showed that 23.8% of BSI, 33.8% of SSI and 71.1% of UTI is caused by Gram- negative Bacilli. (Robert and Jonathan, 2005). Antimicrobial resistance is the developed ability of microorganisms to resist the activity of antibiotics such as antibacterials, antivirals, and so on. Such a threat has evolved due to unnecessary or over use of common antibiotics against diseases. Antimicrobial resistance hence has led to ineffectiveness of some common antibiotics while treating such infections. As a result, infection persists, spreads and even has a good chance of turning into an outbreak. (WHO newsletter no.32, 2017)
Gram-negative bacilli infections have characteristics that require attention. These microorganisms can easily mutate genes that are responsible for the development of mechanisms to show antibiotic resistance in the presence of a selection pressure. Moreover, they have a number of resistance mechanisms that can be differentiated in a vague manner as- those that show multiple mechanisms against the same antibiotics and the other using a single mechanism to affect many antibiotics (Anton and David, 2010). A range of Gram-negative organisms are responsible for hospital- acquired infections, the Enterobacteriaceae Family being the most commonly identified group overall. However, multidrug organisms, including Pseudomonas aeruginosa, Acinetobacter baumanni, and ESBL-producing or Carbapenemase-producing Enterobacteriacea, are increasingly being reported worldwide (Helen et al., 2009). Pseudomonas aeruginosa isGram-negative, rod-shaped, asporogenous, and monoflagellated bacterium. P.aeruginosa grows well at 25°C to 37°C. Pseudomonas areuginosa is a ubiquitous microorganism and has the ability to survive under a variety of environmental conditions. It not only causes disease in plants and animals, but also in patients suffering from severe burns and cystic fibrosis (CF) (Fei et al., 2017). Due to its treatment with a wide range of antibiotics today it has developed into a resistant strain showing multidrug resistance against some common antibiotics. Antibiotic resistance in P. aeruginosa is shown through many cell survival processes such as biofilm formation, efflux pump activity, or in many cases due to resistance phenotype expressing genes. The efflux pumps possessed by Pseudomonas aeruginosa are AmR, which is expressed due to the AmR AB genes.
The RND systems of this microbe also effectively execute resistance through efflux pumps such as MeX AB-OprM, Mex XY-OprM, MexCD-OprJ, MexEF-OprN, MexJK, MexGHI-OpmD, MexVW,MexPQ-OpmE, MexMN, and TriABC (Jürg Dreier, Paolo Ruggerone, et al.,2015) One of which Mex AB-OprM is the most important as it provides intrinsic resistance to a broad spectrum of antibiotics. The genes involved in the synthesis of the Mex AB protein get frequently expressed in the presence of common antibiotics especially β-lactams. (Poole et al., 1993). Research from late 90’s on phytochemistry has brought to light the synergetic activity of natural compounds present in plants with common antibiotics against AMR. (Michael et al., 2007) Further interest in this area has brought the idea of doing in silico research on the phytochemicals present in Indian Medicinal Plants that showed such synergetic activity with the common antibiotics against multidrug resistant strains of Pseudomonas aeruginosa (MDRPA).Furthermore, Pseudomonas aeruginosa is one of the most important causes of Gram-negative infection, especially in immunocompromised individuals and patients undergoing treatment in hospitals. It is the most common pathogen isolated from patients who have been hospitalized for more than 1 week (Marcus et al., 20018). MDRPA has been previously observed to show resistance to a number of antibiotics including Ciprofloxacin, Ceftazidime, Imipenem and Piperacillin during a retrospective study conducted at Beth Israel Deaconess Medical Centre. Some isolates even showed resistance to aminoglycosides such as Gentamicin and Tobramycin. The same study also illustrated that the most frequent sites of Infection were- soft tissue, bone, lungs, UT, all characterised by prolonged infections, multiple and lengthy antipseudomonal antibiotic exposure and long hospitalisation (Laura et al., 20114).
This paper deals with such a hospital isolated strain of Pseudomonas aeruginosa that showed delayed cure, and poor treatment with many antibiotics, isolated at Balaji Dental College and Hospitals, Chennai. The Susceptibility was once again tested with some common antibiotics and the reason for resistance was determined targeting the various mechanisms of resistance.
The perviousresearchers have reported an expanded program on immunization (EPI) activity against Pseudomonas aeruginosa resistant strains. Based on the availability and EPI activity of Wrightia tinctoria was selected as the working sample for this research. Wrightia tinctoria also known as Sweet Indrajao, Dyer’s Oleander, Vetpaalai, belongs to the Apocynaceae family. It is available all over the world and abundantly in parts of South-east Asia mostly Vietnam and India. This tree in South India is used as a Jaundice curative. Apart from this, the plant has been recognized to have a lot of anti-inflammatory properties.Studies have suggested that different parts of the plant are said to possess different phytochemical compounds that play a significant role in the treatment of various bacterial and fungal infections (Gangola et al., 2017; Ekins et al., 2007).
Types of resistance mechanisms that can be shown by a microorganism especially the Gram-negative strain has been illustrated shows in Fig 1.

Fig 1. Various mechanisms of resistance developed by Gram-negative bacterium such as Overexpression of transmembrane efflux pump, mutations in Lipopolysaccharide structure, Loss of porins, β-lactamase production (Anton et al., 2010).
The objective of this research is to study whether the phytochemicals present in Wrightia tinctoria has the efflux pump inhibitory activity against the multidrug resistant strain of Pseudomonas aeruginosa and find out which compound could be responsible for the inhibitory activity of the MexAB-OprM efflux pump shown through in-silico studies.
- Materials and methods
2.1. Virtual visualization
Visualization of the phytochemicals and the proteins that constitute the MexAB pump, in a 3dimensional view, was possible with the help of Biovia Discovery Studio. It’s software that helps create and display the chemical and biomolecules virtually. With Biovia one can research and conduct test to hypothesize prior to conducting wet lab experiments which consumes effort, time and fund. (https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/).
2.2. Ligand identification and Preparation for Screening
A total of 47 phytochemical compounds reported to be present in the plant sample Wrightia tinctoria were selected from literature study. The 3dimensional structures of these phytochemical compounds were retrieved from the database called PubChem. PubChem is a database of chemical compounds that helps us to know achieve the activities of those compounds in biological assays. This database is maintained by the National Centre of Biotechnology Information (NCBI). Structures, IUPAC names, and much other information regarding the chemical compounds under study can be obtained from this database. (https://pubchem.ncbi.nlm.nih.gov/)
The 3dimensional structures of these compounds were achieved from this database and were downloaded in SDF format.
2.3. Receptor structure retrieval
The protein involved in the efflux pump activity in the Pseudomonas aeruginosa was reported to be MexAB-OprM which is expressed due to the OprM genes present in the bacteria (Michael et al., 2007). The structure of this protein complex was achieved from the software named Protein Data Bank. The structure (Pdb id: 1LNW) was downloaded and converted to .mol2 format to be used for molecular docking as our receptor with which all the selected phytochemical compounds present in Wrightia tinctoria would bind as ligands.
2.4. Protein data bank
This is a database that contains three-dimensional structures and other data of complex and large molecules especially proteins and nucleic acids. This database acts as a resource in these areas of biology where structure plays a vital role in the activity under study. (https://www.rcsb.org/)
2.5. Molecular Docking
The MexAB-OprM protein structure along with the ligands, all already converted to .mol2 format were input into docking software named iGEMDOCK.
2.6. iGEMDOCK
iGEMDOCK performs structure based virtual screening and post screening analysis of input which is usually a combination of receptors and ligands and also performs emergent tasks in computer aided drug designing. It acts as an automatic and graphical drug discovery system which does screening, post-analysis and visualization of virtually performed screening where prediction of how chemical molecules bind to a receptor of known three-dimensional structure is executed. (http://gemdock.life.nctu.edu.tw/dock/download/iGEMDOCK-guide.pdf). The docking process was run with 47 ligands and 1 receptor to find out the best poses in post-analysis.
2.6.1. Sample collection
Seeds of Wrightia tinctoria were collected from the forest research department, Vandalur, Chennai. The seeds were shade dried and stored in cool area.
2.6.2. Extract preparation
Seeds of Wrightia tinctoria were crushed with mortar and pestle into a coarse powder and used for extraction with the help of solvent treatment. The following solvents of hexane, ethyl acetate and ethanol were used for extraction. 96.2 g by dry weight of the crushed seeds was taken along with 300 mL of hexane. The vessel (500mL conical flask) was covered with aluminium foil and incubated at 37˚C for 24 hrs. After that incubation filtered by using of Whatman filte No.3. The extraction was noted and stored under the freezer condition. Sediment was air dried and used for the next solvent extraction with ethyl acetate and then ethanol. The 3 extracts of W.tinctoria in hexane, ethyl acetate and ethanol were concentrated by air drying and collected in screw cap vials. The vials were stored at room temperature.
2.6.3. Qualitative phytochemistry
All the three (Hexane, Ethyl acetate, and Ethanol) extracts were diluted to a concentration of 1mg/mL and used for phytochemical screening was done to check for the presence of carbohydrates, tannins, saponins, flavonoids, alkaloids, glycosides, cardiac glycosides, terpenoids, phenol, coumarins, steroids/phyto steroids, phlobatanins, and anthraquinones.
2.6.4. Antibacterial activity
The antibacterial activity of the extracts was checked individually as follows using agar well diffusion method using extract (Ext) and antibiotic-Tetracycline (Tet) at the concentration of 1mg/mL.
| S. No | Sample | Con A | Con B | Con C | Con D | Con E |
| 1. | Plate1 | 10µl Tet | 20µl Ext | 10µl Tet+20µl Ext | 40µl Ext | 10µl Tet+40µl Ext |
| 2. | Plate2 | 10µl Tet | 60µl Ext | 10µl Tet+60µl Ext | 80µl Ext | 10µl Tet+80µl Ext |
| 3. | Plate3 | 10µl Tet | 100µl Ext | 10µl Tet+100µl Ext | 120µl Ext | 10µl Tet+120µl Ext |
The same protocol was followed with the other two extracts as Set2 and 3 respectively.
The three well swabbed LB agar plates with test microorganism were used to puncture 5 wells using a standard metal punch of 0.5 cm diameter, for each extract. The wells were labelled as A, B, C, D and E. the five wells were filled in this manner shows in Table1.
2.6.5. Efflux pump inhibition analysis
The efflux pump inhibition activity of the extracts was tested using Accumulation Assay. 3 test tubes containing 6mL of fresh sterile LB broth was taken. To this, 100µL of Pseudomonas aeruginosa strain broth culture in each tube. 5µl of Tetracycline (1mg/mL) was added to each tube and incubated at 37˚C for 24 hrs. 3mL of culture from each test tube was transferred into 3 micro centrifuge tubes labelled as T1, T2, T3.
T1 was centrifuged at 8000 rpm for 5 minutes at -4˚C to obtain the cell free supernatant. T2 was centrifuged at the same rpm and the pellet was subjected to cell lysis by treating with 100µL of 10% SDS. T3 was also subjected to cell lysis in the same manner as T2. The lysed solution was re-centrifuged and the supernatant was collected. Cell lysed samples were made up to 3mL in the micro centrifuge tubes using Phosphate-buffered saline (PBS). The Tetracycline concentration in each sample was determined spectrophotometrically by reading at (380-450) nm. Reading was taken using LB media with 5µl Tetracycline (1mg/mL) as blank and PBS buffer as blank for the cell lysed samples.
2.6.6. Genomic DNA isolation
Total cell DNA of Pseudomonas aeruginosa was isolated using 1.5mL of the bacterial culture in LB broth. Phenol: Chloroform method of DNA isolation was used to isolate the DNA with the help of differential solubility technique and the isolated DNA was stored in 20µl TE buffer at -20˚C.
2.6.7. Molecular characterisation
The presence of resistive genes in the isolated DNA of the DRPA was identified by performing PCR with the sample DNA using SHV primers as the template. Presence of the genes was checked by gel documentation through agarose gel electrophoresis. 3. Results
3.1. Molecular docking
The phytochemicals that were used for docking are given in the table 2.
MexAB-OprM pump:
PDB ID: 1LNW

Fig 2MexAB OprM efflux pump produced by P. aeruginosa as a mechanism to develop antibiotic resistance.
It was found that the phytochemical Quercetin 3-sophoroside showed in Fig. 1. the best pose with a total energy of -141.931Joules and H-bond value of -45.22 Joules on docking with our receptor under study, MexAB-OprM efflux pump. However, the top 5 compound that showed good posing with our receptor should also be taken into consideration. The five compounds of Quercetin 3-sophoroside (T.E= -141.931J ; H-bond= -41.2182J), Rutin (T.E= -128.26J ; H-bond= -27.84J), 9,12,15- Octadecatrienoic acid (T.E= -120.07J ; H-bond= -17.57J), Quercetin (T.E= -104.62J ; H-bond= -30.56J), Clerosterol (T.E= -104.44J ; H-bond= -10.43J).
Table 2.
| S.No | Compound name | Source | Molecular formula | PubChem ID | IUPAC name |
| 1. | Quercetin 3-sophoroside | Leaf | C27H30O17 | 44259144 | 3-[(2S,5S)-4,5-dihydroxy-6-(hydroxymethyl)-3-[(2S,3R,5S)-3,4,5-trihydroxy-6-(hydroxymethyl) oxan-2-yl] oxyoxan-2-yl] oxy-2-(3,4-dihydroxyphenyl)-5,7-dihydroxychromen-4-one |
| 2. | Kaempferol | Leaf | C15H10O6 |
5280863 | 3,5,7-trihydroxy-2-(4-hydroxyphenyl) chromen-4-one |
| 3. | Quercetin | Leaf | C15H10O7 | 5280343 | 2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxychromen-4-one |
| 4. | Indirubin | Leaf | C16H10N2O2 | 5359405 | (3E)-3-(3-oxo-1H-indol-2-ylidene)-1H-indol-2-one |
| 5. | Indole | Leaf | C8H7N | 798 | 1H-indole |
| 6. | Anthocyanin | Leaf | C15H11O+ | 145858 | 2-phenylchromenylium |
| 7. | Calcones | Leaf | C21H18N2O3S | 6445661 | 4-amino-N-[4-[(E)-3-oxo-3-phenylprop-1-enyl] phenyl] benzene sulfonamide |
| 8. | Catechin | Leaf | C15H14O6 | 9064 | (2R,3S)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol |
| 9. | Isatin | Leaf | C8H5NO2 |
7054 | 1H-indole-2,3-dione |
| 10. | Indigotin | Leaf | C16H10N2O2 | 5318432 | (2E)-2-(3-oxo-1H-indol-2-ylidene)-1H-indol-3-one |
| 11. | Rutin | Leaf | C27H30O16 | 5280805 | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-[[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl] oxymethyl] oxan-2-yl] oxychromen-4-one |
| 12. | Desmosterol | Seed | C27H44O |
439577 | (3S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-[(2R)-6-methylhept-5-en-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol |
| 13. | Clerosterol | Seed | C29H48O |
5283638 | (3S,8S,9S,10R,13R,14S,17R)-17-[(2R,5S)-5-ethyl-6-methylhept-6-en-2-yl]-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol |
| 14. | 9-octadecanoic acid | Seed oil | C19H36O2 |
21677595 | 8-[(1S,2R)-2-octylcyclopropyl] octanoic acid |
| 15. | Alpha-amyrin | Seed oil | C30H50O |
73170 | (3S,4aR,6aR,6bS,8aR,11R,12S,12aR,14aR,14bR)-4,4,6a,6b,8a,11,12,14b-octamethyl-2,3,4a,5,6,7,8,9,10,11,12,12a,14,14a-tetradecahydro-1H-picen-3-ol |
| 16. | Squalene | Seed oil | C30H50 |
638072 | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene |
| 17. | Gamma-tocopherol | Seed oil | C28H48O2 | 92729 | (2R)-2,7,8-trimethyl-2-[(4R,8R)-4,8,12-trimethyltridecyl]-3,4-dihydrochromen-6-ol |
| 18. | Campesterol | Seed oil | C28H48O | 173183 | (3S,8S,9S,10R,13R,14S,17R)-17-[(2R,5R)-5,6-dimethylheptan-2-yl]-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol |
| 19. | Betulin | Seed oil | C30H50O2 | 72326 | (1R,3aS,5aR,5bR,7aR,9S,11aR,11bR,13aR,13bR)-3a-(hydroxymethyl)-5a,5b,8,8,11a-pentamethyl-1-prop-1-en-2-yl-1,2,3,4,5,6,7,7a,9,10,11,11b,12,13,13a,13b-hexadecahydrocyclopenta[a]chrysen-9-ol |
| 20. | Lupenone | Seed oil | C30H48O |
92158 | (1R,3aR,5aR,5bR,7aR,11aR,11bR,13aR,13bR)-3a,5a,5b,8,8,11a-hexamethyl-1-prop-1-en-2-yl-2,3,4,5,6,7,7a,10,11,11b,12,13,13a,13b-tetradecahydro-1H-cyclopenta[a]chrysen-9-one |
| 21. | Phytol | Seed oil | C20H40O | 5280435 | (E,7R,11R)-3,7,11,15-tetramethylhexadec-2-en-1-ol |
| 22. | Pentadecanoic acid, ethyl ester | Flower | C17H34O2 | 38762 | ethyl pentadecanoate |
| 23. | Dimethyl phthalate | Flower | C10H10O4 | 8554 | dimethyl benzene-1,2-dicarboxylate |
| 24. | Hydroquinone | Leaf EO | C6H6O2 | 785 | benzene-1,4-diol |
| 25. | 9,12,15-octadecatrienoic acid | Leaf EO | C28H34O4 | 57397401 | (5,8-dioxonaphthalen-1-yl)
(9Z,12Z,15Z)-octadeca-9,12,15-trienoate |
| 26. | 2-heptenal | Seed oil | C7H12O | 5283316 | (E)-hept-2-enal |
| 27. | Phenol | Leaf EO | C6H6O | 996 | Phenol |
| 28. | Benzyl alcohol | Leaf EO | C7H8O | 244 | phenyl methanol |
| 29. | n-methyl-n-2-pyridinyl formamide | Leaf EO | C7H8N2O | 572611 | N-methyl-N-pyridin-2-ylformamide |
| 30. | 2-methoxy phenol | Leaf EO | C7H8O2 | 460 | 2-methoxyphenol |
| 31. | 2,4-dimethyl phenol | Leaf EO | C8H10O | 7771 | 2,4-dimethylphenol |
| 32. | 4-ethyl phenol | Leaf EO | C8H10O |
31242 | 4-ethylphenol |
| 33. | 3,5-dichloro phenol | Leaf EO | C6H4Cl2O | 11571 | 3,5-dichlorophenol |
| 34. | 2,3-dimethyl phenol | Leaf EO | C8H10O | 10687 | 2,3-dimethylphenol |
| 35. | n-hexadecenoic acid | Leaf EO | C16H32O2 | 985 | hexadecenoic acid |
| 36. | 2-methoxy 4-methylphenol | Leaf EO | C8H10O2 | 7144 | 2-methoxy-4-methylphenol |
| 37. | Alpha-cubebene | Leaf EO | C15H24 | 86609 | |
| 38. | Alpha-caryophyllene | Leaf EO | C15H24 |
5281520 | (1E,4E,8E)-2,6,6,9-tetramethylcycloundeca-1,4,8-triene |
| 39. | Butylated hydroxyl toluene | Leaf EO | C15H24O | 31404 | 2,6-ditert-butyl-4-methylphenol |
| 40. | Dodecanoic acid | Leaf EO | C12H24O2 | 3893 | dodecanoic acid |
| 41. | Caryophyllene oxide | Leaf EO | C15H24O |
1742210 | |
| 42. | 1-ethyl heptyl-benzene | Leaf EO | C13H20 |
14115 | heptyl benzene |
| 43. | 1-pentyl heptyl-benzene | Leaf EO | C18H30 |
17629 | dodecan-6-ylbenzene |
| 44. | 3-ethyldecylbenzene | Leaf EO | C18H30 |
56994984 | 3-ethyldecylbenzene |
| 45. | Coumarin | Seed | C9H6O2 | 323 | chromen-2-one |
| 46. | Anthraquinone | Seed | C14H8O2 | 6780 | anthracene-9,10-dione |
| 47. | 4-methylphenol | Leaf EO | C7H8O | 2879 | 4-methylphenol |



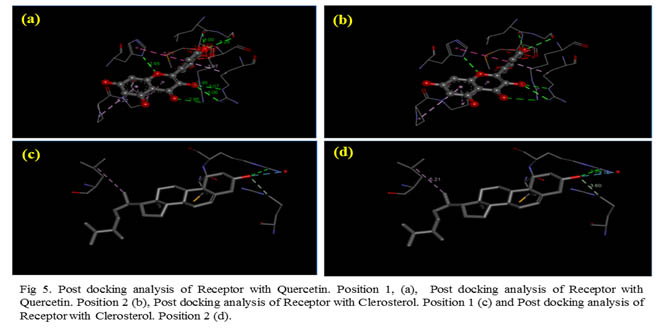
Out of the top five best poses Quercetin 3-sophoroside showed the best fit with a bond energy of -141.931Joules.
3.2. Microbiological & phytochemical assays
The Wrightia tinctoria seeds that resembled paddy grains in colour produced a light scarlet red solution on extraction with different solvents. The extracts were observed to turn darker in colour as the extracts concentrated on air drying. The antibiogram showed Fig4 a, that the P. aeruginosa isolated was resistant to methicillin and rifampicin but not to ciprofloxacin and streptomycin.
3.3. Antibiotic susceptibility test
The well diffusion assay done to check for antibacterial activity showed zones of inhibition around wells where tetracycline was used alone and around wells where it was along with the extract.
Zones were observed in wells which had 10µl Tet, (20µl Ext + 10µlTet), (40µl Ext + 10µl Tet), (60µL Ext +10µl Tet), (80µl Ext +10µl Tet), (100µl Ext + 10µl Tet), (120µl Ext + 10µl Tet).

Fig. 6 Antibiogram of Wrightia tinctoria seeds extract (a), and (b) phytochemical assays.
These observations were recorded while working with the hexane extract of W.tinctoria. The zones observed around well which had both the antibiotic and the extract however, showed larger diameter than with only the antibiotic (Tet). The diameter difference observed was (0.5-1 cm). This may be considered as positive synergistic activity of the plant extracts.
3.4. Accumulation assay
Table 4. The spectrophotometric observation of samples from the accumulation assay furnished the results listed in the table 4.
| Nanometres | Sample 1 | Sample2 | Sample3 |
| 380 | 0.0124 | -0.1487 | -0.0069 |
| 390 | 0.0117 | -0.1496 | -0.0055 |
| 400 | 0.0124 | -0.1527 | -0.0062 |
| 410 | 0.0132 | -0.1527 | -0.0062 |
| 420 | 0.0139 | -0.1520 | -0.0062 |
| 430 | 0.0145 | -0.1527 | -0.0056 |
| 440 | 0.0139 | -0.1520 | -0.0060 |
| 450 | 0.0143 | -0.1525 | -0.0060 |
The spectrophotometer readings of absorbance from (380-450) nm show no accumulation of drug in samples 2 and 3. Thus, no drug accumulation indicated absence of an active efflux pump mechanism.
3.5. Polymerase chain reaction
The isolated DNA of the resistant Pseudomonas aeruginosa was taken for PCR to check for the presence of resistive genes. PCR was done with SHV primer. The PCR product when run through Agarose showed in Fig. 7 amplification. Presence of SHV sequences that belong to the ESBL family was confirmed which also confirms the presence of resistive genes that were capable of producing antibiotic destroying enzymes.

Fig. 7 Agarose gel electrophoresis done post PCR showing the amplification of the SHV gene
- Discussion
In silico analysis was done to find out what possible compounds extracted from natural sources could be used to block the MexAB efflux pump activity shown by the MDR strain of Pseudomonas aeruginosa. In silico methods furnished a speedy reporting of such compounds that showed best posing using the method of Molecular Docking. Wet lab assays would have instead taken much more time to complete and conclude with precise results. The receptor that was chosen for study, the MexAB efflux pump, was reported to show multidrug resistance against β-lactams,fluroquinolones, novobicin, chloramphenicol, macrolides, sulfonamides, trimethoprims, tetracycline and some other detergents and dyes (David et al., 2002). The ligands that showed best interaction with the receptor under study are quercetin 3-sophoroside, rutin, 9,12,15-octadecatrienoic acid, quercetin, clerosterol, and gamma-tocopherol, in the order of increasing binding energy. All these five compounds have been reported to have medicinally important value. Abdollahzadeh et al., 2011, reported that quercetin 3-sophoroside isolated from Korean Papaver rhoeas bee pollen showed neurominidase inhibitory activities. A combination of quercetin and rutin has been reported to enhance antibacterial activity of flavonoids, against Bacillus cereus and Salmonella enteritidis (Hidetoshi et al., 2002). In general, quercetin glycosides isolated from red fruits showed antimicrobial, antiviral activity. (Khatereh and Reza, 2013). A report on phytochemical screening of Punica granatum pomegranate showed that compounds like gamma-tocopherol showed antibacterial activity against many oral pathogens including Staphylococcus aureus, Streptococcus mutans, Streptococcus salivarius, Staphlyococcus epidermis, Lactobacillus acidophilus and also antifungal activity against Candida albicans (Nord et al., 972) Even the compound 9,12,15- octadecatrienoic acid isolated from Aloe Vera succulent, showed significant antibacterial and antifungal activity against Staphylococcus aureus, Streptococcus pyogens, Pseudomonas aeruginosa and E.coli. it showed inhibition of fungus such as Aspergillus flavans and Aspergillus niger. (Arunkumar and Muthuselvam, 2009). Sterols such as Clerosterol which also showed good binding energy with the MexAB efflux pump system has been reported to have in vitro antimicrobial activity against fungi, yeast, and bacteria using ethanol and aqueous extracts of Cassia fistula. It also has been reported to show anti-inflammatory activity. (Patrícia et al., 2007)
A recent homology on the RND MexXY another overexpressed protein present in MDR Pseudomonas aeruginosa showed insilico screening with common antibiotics. The study showed that the efflux pump exhibited an interaction as follows Penicillin (T.E= -8.3J), Chloramphenicol (T.E= -6.7J), Lincomycin (T.E= -7.2J) and Tetracycline (T.E= -8.2). This shows that phytochemicals from natural sources act as extremely good efflux inhibitors than synthetic drugs on comparing their bonding energies (Vijaya et al., 2012). A review on the extrusion pump MexAB-OprM in Pseudomonas aeruginosa proposes that the pump itself could be used as a high throughput screening system for the designing of new novel antimicrobials (Kazuki, et al., 2015). On performing the Antibiogram, it was observed that the given strain showed resistance to Methicillin and Rifampicin. Targeting of the efflux pump mechanism as a possible reason for resistance shown by Pseudomonas aeruginosa using extracts of Wrightia tinctoria as a potential efflux pump inhibitor was executed (Mahendra et al., 2014). Accumulation assay, which was done to check the accumulation of the drug after blocking efflux pumps with the plant extract showed no accumulation. (Li et al., 1994) it could either be interpreted that the extract had no activity or else there is no active efflux pump mechanism of resistance. But, the tests done to check for antibacterial activity showed positive inhibitory effect and moreover W.tinctoria has also been reported to show positive EPI activity (Mahendra and Nityanand, 2009).
Hence, it could either possible that there was no active efflux pump in the isolated strain. Moreover, it is reported that an active efflux pump mechanism in P.aeruginosa PA01, wild type, could naturally resist Aminoglycosides. A mutation in the genes has been reported to be the reason for the loss of the active efflux mechanism. (Julio et al., 1999; In-Kyoung et al., 2016) Our antibiogram has worked with Streptomycin which also confers no resistance being an Aminoglycoside. The study was therefore, taken to molecular precision. The target of the investigation now was to detect the presence of resistive genes. Polymerase Chain Reactions of the DNA from the isolated sample was done with SHV primers which showed amplification confirming the presence of the SHV gene sequence that relate to the ESBL family (Dirk et al., 2009). Resistance to drugs such as Methicillin and Oxacillin in Staphylococcus aureus is determined by the presence of SCCmec, a mobile genetic element present in their chromosomes which contains the mecA resistance gene. (John W. Pearman, Fred C. Tenover, et al, 2002) Rifampicin resistance in M.tuberculosis and E.coli has been reported to be due to mutation in the gene encoding foe RNA polymerase subunitβ (rpoB) (Telenti et al, 1993) It is described as a “single step” high level resistance pattern that arises due to missense mutations in the 69-bp region of rpoB. (Kapur et al., 1994)
Resistance to drugs such as Methicillin and Oxacillin in Staphylococcus aureus is determined by the presence of SCCmec, a mobile genetic element present in their chromosomes which contains the mecA resistance gene. (Barben and Schmid, 2008) Rifampicin resistance in M.tuberculosis and E.coli has been reported to be due to mutation in the gene encoding foe RNA polymerase subunitβ (rpoB). (Telenti et al, 1993) It is described as a “single step” high level resistance pattern that arises due to missense mutations in the 69-bp region of rpoB (Kapur et al., 1994). Modern quinolones such as Ciprofloxacin show increased activity and less frequency of being resisted by bacterial resistance mechanisms. The target of these drugs is previously reported to be a subunit of the bacterial enzyme DNA gyrase and also another site in addition to this. It is also reported that this additional target decreases the susceptibility of the organism to Rifampin (Hooper et al., 1987). This phenomenon could also be a possible reason for the resistance shown by the P.aeruginosa under study against Rifampicin as seen in our Antibiogram. The study concludes that Pseudomonas aeruginosa that is responsible for most superficial infections has now developed resistance against many common antibiotics including Methicillin and Rifampicin. Presence of SHV genes being one of the reasons of resistance which is capable of showing activity like most ESBL organisms. Further study and molecular characterization of the strain is needed to find out any other potential mechanisms of resistance present in MDRPA and a cure from natural sources to fight against the crisis of Antibiotic Resistance in Nosocomial Infections.
On performing the Antibiogram, it was observed that the given strain showed resistance to Methicillin and Rifampicin. Targeting of the efflux pump mechanism as a possible reason for resistance shown by Pseudomonas aeruginosa using extracts of Wrightia tinctoria as a potential efflux pump inhibitor was executed. (Mahendra et al., 2014) Accumulation assay, which was done to check the accumulation of the drug after blocking efflux pumps with the plant extract showed no accumulation. (Li et al., 1994) Therefore, it can either be interpreted that the extract had no activity or else there is no active efflux pump mechanism of resistance. But, the tests done to check for antibacterial activity showed positive inhibitory effect and moreover W.tinctoria has also been reported to show positive EPI activity (Mahendra and Nityanand 2009). A mutation in the genes has been reported to be the reason for the loss of the active efflux mechanism. Our antibiogram has worked with Streptomycin which also confers no resistance being an Aminoglycoside. The study was therefore, taken to molecular precision. The target of the investigation now was to detect the presence of resistive genes. Polymerase Chain Reactions of the DNA from the isolated sample was done with SHV primers which showed amplification confirming the presence of the SHV gene sequence that relate to the ESBL family (Dirk et al., 2010).
Resistance to drugs such as Methicillin and Oxacillin in Staphylococcus aureus is determined by the presence of SCCmec, a mobile genetic element present in their chromosomes which contains the mecA resistance gene. (Keiko et al., 2002) Rifampicin resistance in M.tuberculosis and E.coli has been reported to be due to mutation in the gene encoding foe RNA polymerase subunitβ (rpoB). It is described as a “single step” high level resistance pattern that arises due to missense mutations in the 69-bp region of rpoB (Kapur et al., 1994). Modern quinolones such as Ciprofloxacin show increased activity and less frequency of being resisted by bacterial resistance mechanisms. The target of these drugs is previously reported to be a subunit of the bacterial enzyme DNA gyrase and also another site in addition to this. It is also been reported that this additional target decreases the susceptibility of the organism to Rifampin (Hooperet al., 1987). This phenomenon could also be a possible reason for the resistance shown by the P.aeruginosa under study against Rifampicin as seen in our Antibiogram. Thus, from this study we can hypothesize and conclude that the extracts of Wrightia tinctoria possess such valuable phytochemical compounds might be used as an inhibitor alone or as a synergistic inhibitor with common antibiotics. However, further wet lab study and much more precise results are required in this area to validate the study.
- Conclusion
The study concludes that Pseudomonas aeruginosa that is responsible for most superficial infections has now developed resistance against many common antibiotics including methicillin and rifampicin. Presence of SHV genes being one of the reasons of resistance which is capable of showing activity like most ESBL organisms. Further study and molecular characterization of the strain is needed to find out any other potential mechanisms of resistance present in MDRPA and a cure from natural sources to fight against the crisis of antibiotic resistance in nosocomial infections.
Funding: The authors received no specific funding for this work.
Conflicts of Interest: None
References
- Abdollahzadeh, S.H., Mashouf, R.Y., Mortazavi, H., Moghaddam, M.H., Roozbahani, N., and Vahedi, M., 2011. Antibacterial and Antifungal Activities of Punica Granatum Peel Extracts Against Oral Pathogens. J Dent (Tehran), 8,1-6.
- Arunkumar, S., Muthuselvam, M., 2009. Analysis of Phytochemical Constituents and Antimicrobial Activities of Aloe vera L. Against Clinical Pathogens. World Journal of Agricultural Sciences 5, 572-576.
- Anton, Y.P and David, C.H., 2010. Hospital-Acquired Infections Due to Gram-Negative Bacteria. N Engl J Med, 362, 11804-183.
- Barben, J and Schmid, J., 2008. Dental units as infection sources of Pseudomonas aeruginosa. Eur. Respir. J, 32, 1122-1123.
- Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas.
- Dirk, M.L., Verena, G., Maya, R., Jan, W., Klaus, S., Thomas, A.W., Cornelius, K., Rolf, D.S., Till, T.B., 2010. Integrated detection of extended-spectrum-beta-lactam resistance by DNA microarray-based genotyping of TEM, SHV, and CTX-M genes. J Clin Microbiol, 48, 460-71.
- David, M.L., 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis, 34, 634-40.
- Ekins, S., Mestres, J., Testa, B., 2007. In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling Br J Pharmacol, 152, 9-20.
- Fei, C., Gukui, C., Yiwei, L., Yongxin, J., Zhihui, C., Yang, L., Liang, Y., Shouguang, J., and Weihui, Wu., 2017. Pseudomonas aeruginosa Oligoribonuclease Contributes to Tolerance to Ciprofloxacin by Regulating Pyocin Biosynthesis. Antimicrob Agents Chemother. 2017 61, e02256-16.
- Gangola, S., Khati, P., Bhatt, P., Parul and Anita, S., 2017. India as the Heritage of Medicinal Plant and their Use. Curr Trends Biomedical Eng & Biosci 4, 0050.
- Helen, W.B., George, H.T., John, S.B., John, E.E., David, G., Louis, B.R., Michael, S., Brad, S., John, B., 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis, 48, 1-12.
- Hidetoshi, A., Hitoshi, A., Gen-ichi, D., 2002. Rutin-enhanced antibacterial activities of flavonoids against Bacillus cereus and Salmonella enteritidis. Biosci Biotechnol Biochem, 66, 1009-14.
- Hooper, D.C., Wolfson, J.S., Ng, E.Y., Swartz, M.N., 1987. Mechanisms of action of and resistance to ciprofloxacin. Am J Med, 27, 12-20.
- In-Kyoung., Byung, S.H., Dae-Won, K., Ji-Yul, K., E-Eum, W., Yoon-Ju, L., Hwa, J.C., Bong-Sik, Y., 2016. Characterization of Neuraminidase Inhibitors in Korean Papaver rhoeas Bee Pollen Contributing to Anti-Influenza Activities In Vitro. Planta Med, 82, 524-529.
- Julio, R.A., Thilo, K.H., Hiroshi, N., And Patrick, P., 1999. Involvement of an Active Efflux System in the Natural Resistance of Pseudomonas aeruginosa to Aminoglycosides. Antimicrob. Agents Chemother, 2624–2628.
- Jürg, D., and Paolo, R., 2015. Interaction of antibacterial compounds with RND efflux pumps in Pseudomonas aeruginosa. Front. Microbiol. 6, 660.
- Kapur, V., Li, L.L., Iordanescu, S., Hamrick, M.R., Wanger, A., Kreiswirth, B.N., and Musser, J.M., 1994. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 32, 1095-1098.
- Kazuki, S., Kenichi, U., Keiji, A., Yoshimi, M., Hideaki, M., Tasuke, A., Emiko, I., Taiji, N., and Hiroshi, Y., 2015. Development of a Novel Antimicrobial Screening System Targeting the Pyoverdine-Mediated Iron Acquisition System and Xenobiotic Efflux Pumps. Molecules, 20, 7790–7806.
- Keiko, O., Kozue, I., John, D.T., Warren, B., Jan, M.B., Frances, G. O.B., Geoffrey, W. C., John W.P., Fred, C.T., Maria, K., Chuntima, T., Teruyo, I., and Keiichi, H., 2002. Dissemination of New Methicillin-Resistant Staphylococcus aureus Clones in the Community. J Clin Microbiol, 40, 4289-4294.
- Khatereh, K., Reza, H., 2013. Determination and comparison of major polyphenol of four red fruits using high performance liquid chromatography (hplc) with diode-array detection. Journal of Microbiology, Biotechnology, and Food Sciences.3, 250-252.
- Laura, F., Susanna, L., Michaela, C., Julia, B., Giuseppe, P., Marta, L.C.D.A., Chiara, B., Barbara, L., Stefania, R., Edoardo, C., Maurizio, M., Mike, S., Paola, B., 2014. Epidemiology and clinical outcomes of multidrug-resistant, gram-negative bloodstream infections in a European tertiary pediatric hospital during a 12-month period. Pediatr infect Dis J, 33,929-32.
- Li, X.Z., Livermore, D.M., and Nikaido, H., 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother, 1732-174.
- Mahendra S. K., and Nityanand, P.V., 2014. Wrightia tinctoria R. Br.-a review on its ethnobotany, pharmacognosy and pharmacological profile. Journal of Coastal Life Medicine, 10, 826-840.
- Mahendra S.K., and Nityanand, P.V., 2009. Antibacterial evaluation and phytochemical analysis of wrightia tinctoria (roxb.) R. Br. Leaves. Pharmacologyonline 2: 808-813.
- Marcus, F., Burke, A.C., Klaus, D.L., Kevin, G.G.L., 2018. Pseudomonas aeruginosa Medscape.
- Marilee, D. O., Douglas, N.F., Robert, M.L., Rose, J., 2005. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherpy, 25, 1353-64.
- Michael, S., Laura, Piddock, J.V., Simon, G., 2007. Bacterial efflux pump inhibitors from natural sources. J Antimicrob Chemother, 59, 1247-60.
- Moorthy, K., Aparna, A., Punitha, T., Vinodhini, R., Suresh, M., And Thajuddin, N., 2012. In vitro screening of antimicrobial activity of wrightia tinctoria (roxb.) R. Br. Asian J Pharm Clin Res, 5, 54-58
- Nord, C.E., Sjöberg, L., & Wadström, T., 1972. Pseudomonas Aeruginosa in Oral Infections. Acta Odontol. Scand, 30, 371-381.
- Patrícia, S., Samanta, P.A., Márcia, S.C.M., Frederico, O.P., André, G.T., 2007. Isolation of antileishmanial sterol from the fruits of Cassia fistula using bioguided fractionation, 21,644-647.
- Poole, K., Krebes, K., McNally, C., and Neshat, S., 1993. Multiple antibiotic resistances in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 175, 7363–7372.
- Robert, G., Jonathan, R. E., 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis, 41, 848-54.
- Telenti, A., Imboden, P., Marchesia, F., Matter, L., Schopfer, K., Bodme, T., Lowrie, D., Colston, M.J., Cole, S., 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis, The lancet, 341, 647-651.
Vijaya, B.B., Munichandrababu, T., Bhuvaneswar, C., Rajendra, W., 2012. Homology And Molecular Docking Of Pseudomonas Aeruginosa Rnd Mexy Efflux Pump. Online journal of bioinformatics, 13, 274-293.
